Current Pediatric Research
International Journal of Pediatrics
Next-generation sequencing identified a novel PLP1 deletion in a proband with connatal Pelizaeus-Merzbacher disease.
Giovanni Savarese1, Francesca Felicia Operto2, Raffaella Ruggiero1, Alfonso Ciriello3, Luigia De Falco1, Roberta Mazza2, Fausta Piscopo3, Alberto Verrotti4, Giangennaro Coppola5, Alessandro Frolli3*
1AMES Polidiagnostic Instrumental Centre of Casalnuovo, Italy.
2Child Neuropsychiatry Unit, Department of Basic Medical Sciences, Neuroscience and Sense Organs, University of Bari “Aldo Moro”, Italy.
3Disability Research Center, International University of Rome, Italy.
4Child Neuropsychiatry Unit, Department of Basic Medical Sciences, Neuroscience and Sense Organs, University of Salerno, Italy.
5Department of Pediatrics, University of L'Aquila, Italy.
- Corresponding Author:
- Alessandro Frolli
Disability Research Center, International
University of Rome, Via Cristoforo Colombo
200, Rome, Italy
Tel: +39 3474910178
Email: alessandro.frolli@unint.eu
Accepted date: October 23rd, 2018
Pelizaeus-Merzbacher disease (PMD; MIM #312080) is a rare X-linked recessive disorder characterized with early nystagmus, hypotonia, ataxia and neurodegeneration. PMD is caused by mutations in PLP1 gene that encodes for the myelin protein Proteolipid Protein 1 (PLP1) and the spliced variant DM20. Generally, patients with PLP1 missense mutations show the most severe form of PMD (connatal form), whereas PLP1 duplications, that are the most common mutations result in the classical PMD. Deletions and null mutations are associated to the mild form of PMD and Hereditary Spastic Paraplegia type 2 (SPG2). The present paper shows a boy of 17-year-old with clinical diagnosis of PMD in which we found a novel PLP1 deletion using Next Generation Sequencing (NGS). Clinical exome sequencing was performed on the affected proband as well as his mother. Libraries were generated according to manufacturer’s protocols using TruSight One kits (Illumina Inc., San Diego, CA, USA) and carried out on NEXT Seq 500 (Illumina Inc., San Diego, CA, USA) to mean sequencing depth of at least 100x. Sequencing results suggested of a complete or partial deletion of PLP1 gene. MLPA (Multiplex Ligation-dependent Probe Amplification) analysis confirmed a deletion in exon 6 of PLP1 inherited from asymptomatic carrier mother. In conclusions, although the patient we described had a severe but stable phenotype, even if he present a deletion in PLP1, suggesting that also deletions give rise to variable phentotypes of PMD. This report also extends the spectrum of PLP1 mutations and highlights the diagnostic utility of NGS to investigate complex clinical phenotypes.
Keywords
Pelizaeus-Merzbacher disease, Next Generation Sequencing (NGS), Dysmyelination, PLP1 deletion.
Introduction
Next-Generation sequencing has been increasingly implemented in clinical diagnostics and has rapidly become a well-established technology In addition to whole genome/exome sequencing, large gene panels of clinically associated genes are used as high-throughput sequencing tests in many clinical centers [1,2]. A diagnostic yields based on some disease-specific study groups may be significantly higher than overall diagnostic yields of a large unselected diagnostic cohort [3].
Pelizaeus–Merzbacher disease (PMD MIM 312080) is a rare dysmyelinating X-linked disorder affecting the central nervous system, with lesions in the cerebral and spinal cord white matter. Most clinical findings are hypotonia, nystagmus and delayed development of motor skills already present in the early childhood [4]. The clinical spectrum ranges from severe connatal cases to relatively benign adult forms [5]. PMD usually concern mutations in the Xq21-22 locus of the PLP1 (MIM# 300401) gene that encodes the main component of myelin in the Central Nervous System (CNS), a four-pass transmembrane protein called proteolipid protein 1 (PLP1) [6]. It seems that abnormal PLP1 impairs protein trafficking and induce apoptosis in oligodendroglia. Consequently PLP1 deficiency is related to an insufficient generation of myelin sheaths with the remaining proteins [5]. In literature different types of PLP1 mutations are reported, such as duplication (60-70%), missense mutation/point mutations (15-20%), insertions and deletions [2,7]. We herein report a case of a young patient with clinical diagnosis of PMD in which we found a novel PLP1 mutation, exon 6 deletion, through a combination of NGS and MLPA analyses.
Materials and Methods
Patient
A 17 year’s old boy was referred to our attention becauseof a clinical diagnosis of PMD. At the same time the proband’s pregnant mother was investigated. Clinical information including family history, birth history, developmental history, physical findings, motor and cognitive functions, laboratory findings, brain images and treatment history were collected after receiving their written informed consent.
Molecular diagnosis
DNA Extraction
Genomic DNA was isolated from blood samples of the parents the proband, proband’s aunt, and of amniotic fluid using the QIAamp DNA blood Mini Kit (QIAGEN) following the manufacturer’s protocol.
Clinical exome sequencing
Clinical Exome Sequencing (ES) was carried out on the affected proband as well as his mother and amniotic fluid. Libraries were generated according to manufacturer’s protocols using TruSight One kits (Illumina Inc., San Diego, CA, USA). Sequencing was carried out on NEXT Seq 500 (Illumina Inc., San Diego, CA, USA) to mean sequencing depth of at least 100x. Exome data processing, variant calling, and variant annotation were performed as described in Supplementary materials and methods. We only selected variants affecting coding exons or canonical splice sites. Subsequently, synonymous variants were filtered out, and only rare variants (frequency of <0.1% in both dbSNP138 and our in-house database containing >1,000 exomes) with high quality were reported.
MLPA (Multiple Ligation-dependent Probe Amplification reaction)
PLP1 dosage analysis was performed in the proband, his parents, his aunt (twin sister of proband’s mother) and amniotic fluid by Multiple Ligation-dependent Probe Amplification reaction (MLPA). The test used SALSA P022 B2 (MRC-Holland), containing probes covering all PLP1 exons and 20 control probes located on the flanking regions of PLP1 gene on the X chromosome. Reactions were performed according to the manufacturer’s recommendations (100 ng of DNA/reaction, hybridization time 16 h and 35 PCR cycles). Reaction products were separated with 3130 Genetic Analyzer (Applied Biosystems). The analysis was performed using Coffalyser. NET software (by MRC-Holland) (standard parameters, dosage ratio boundaries <0.75 and >1.25 for deletion and duplication respectively).
PCR analysis
All coding exons and exon–intron boundaries of PLP1 were amplified using primers designed with Primer3web version 4.1.0. Primers sequences are available on request. The genomic sequence from GenBank accession numbers NM_001128834 was used as reference sequence. The amplified products were isolated by electrophoresis on 1% agarose gel and purified using the QIAamp purification kit (Qiagen, Valencia, CA).
Results
Clinical reports
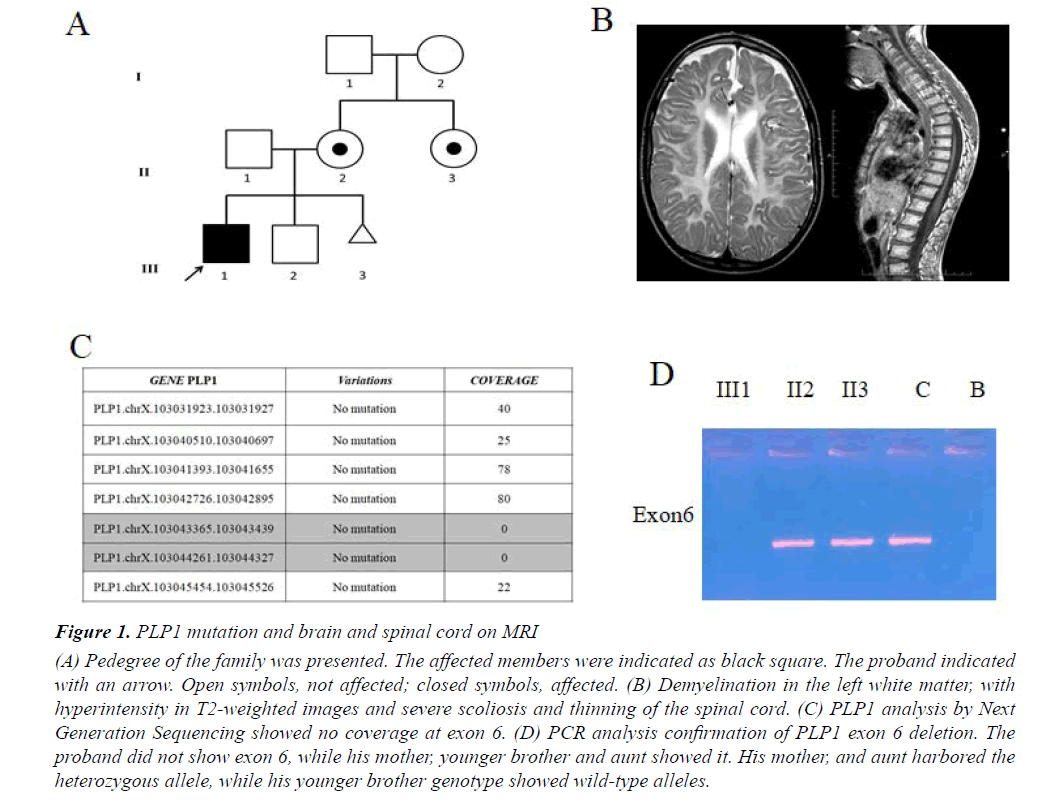
We report a case of 17-year-old male, born at term after an uneventful pregnancy from healthy, unrelated parents, in the absence of a family history of neurological illness (Figure 1A). Body weight at birth was 2750 g. Since the first months of life, the child was hypotonic and unable to maintain the head upright; he presented spontaneous nystagmus in all directions and microcephaly. Towards the end of the first year of life, he showed global psychomotor delay, generalized hypotonia and muscle hypotrophy, peripheral limb hypertonia and spontaneous nystagmus. During this period the diagnosis of PMD was suspected, but molecular analysis performed with classical methods excluded PLP1 duplication. At the age of 4 years, decreased motor conduction velocity with small compound muscle action potential amplitude suggested a sensorimotor polyneuropathy. A showed-down electric activity on frontal areas bilaterally was detected byEEG in sleep. A brain MRI showed supratentorial, symmetrical abnormal white matter with hyperintensity in T2-weighted images, thus suggesting demyelination. EMG showed myogenic changes and also visual evoked potentials were altered. The patient also performed muscle biopsy showing congenital muscular dystrophy. Metabolic tests were normal. At age of 14 years, the patient was severely unable to swallow due to the paucity of tongue movements, , as shown by an X-ray. One year later an eye examination showed visual impairment. Brain and cord MRI displayed demyelination in the left white matter with hyperintensity in T2-weighted images, severe scoliosis and thinning of the spinal cord (Figure 1B). An sleep EEG showed isolated spikes on the right occipital areas. Currently, the boy is affected by severe intellectual disability, spastic quadriplegia, aphasia, irregular sleep-wake cycle that has benefited from low daily doses of periciazine.
Figure 1: PLP1 mutation and brain and spinal cord on MRI
(A) Pedegree of the family was presented. The affected members were indicated as black square. The proband indicated
with an arrow. Open symbols, not affected; closed symbols, affected. (B) Demyelination in the left white matter, with
hyperintensity in T2-weighted images and severe scoliosis and thinning of the spinal cord. (C) PLP1 analysis by Next
Generation Sequencing showed no coverage at exon 6. (D) PCR analysis confirmation of PLP1 exon 6 deletion. The
proband did not show exon 6, while his mother, younger brother and aunt showed it. His mother, and aunt harbored the
heterozygous allele, while his younger brother genotype showed wild-type alleles.
Molecular studies
Starting from aprevious molecular diagnosis which ruled out PLP1 duplication we performed clinical Exome Sequencing (ES) in order to investigate point mutations in PLP1 as well as variations in other associated PDM genes or possible candidate genes. Sequencing analysis on the proband and amniotic fluid not revealed a pathogenic mutation in PLP1 as well as in the other genes causing the other forms of hypomyelinating leukodystrophy of PMD. However in our patient a proportion of PLP1 gene showed sequencing depth of 0 (Figure 1C). We confirmed a deletion in exon 6 in hemizygous state of PLP1 gene by MLPA analysis. According to MLPA, the subject had a diagnostic index equal to 0 for exon 6 inherited from asymptomatic carrier mother (Figure 1A). PCR analysis of PLP1 gene confirmed the deletion of exon 6 only in the proband as shown in Figure 1D. We also found the same deletion in his aunt in heterozygous state, whereas no mutation was found in his father and his younger brother (Figure 1A). Finally, molecular analysis performed on amniotic fluid did not reveal the same deletion (Figure 1A).
Discussion
Different PLP1 mutations give rise to clinical phenotype of PMD ranging from most severe connatal form, with nystagmus, seizures and sever motor deficits, and death which occur during the infancy, to relatively mild SPG2 and PLP1 null syndrome associated to peripheral neuropathy [8].
PLP1 gene duplications are the most frequent mutation in PMD (60-70%) and generally results in the classical form of the disease [6,9,10]. In the 15-20% of patients point mutations, including missense, nonsense, and splicing have been detected, while null mutations are rare [11-14]. Also insertions [15], deletions [16] and translocations [17] have been described as responsible for PMD. In general, missense mutations results to more severe forms of the disease, deletions and null mutations to mild PMD and SPG2, whereas the most common variations, duplications, result in the classical-intermediate form of PMD. In this study, we described a PMD patient with a novel mutation in PLP1 gene, which cause a deletion of entire exon 6. To date, this mutation has not been reported in any of the publicly databases (ExAC, ESP, 1000G) or in literature. The child showed hypotonia with failure to keep the head straight, nystagmus and microcephaly. Later he showed progressive psychomotor delay, muscle hypotrophy and peripheral limb hypertonia up to a severe intellectual disability, spastic quadriplegia, aphasia and irregular sleep-wake cycle.
Complete PLP1 gene deletions are very rare (approximately 2% of patients) and only few studies described clinical and molecular characterization of these patients [17,18]. The first report described a fourgeneration family with two living affected male with a deletion of PLP1 gene encompassing the promoter region, the entire coding region, and the 3' untranslated region spanning at least 29 kb of genomic DNA [18]. Additional cases with complete or partial PLP1 deletion clarified the possible molecular mechanisms underlying these genomic rearrangement as well as brain MRI findings [16,19-21]. Up to now few affected females with X-linked PMD have been described with the development of neurodegenerative symptoms later in life [22-24]. Brender et al. described the first affected female with a PLP1 deletion and a severe but stable phenotype, probably due a second event of skewed X chromosome inactivation [25]. Only one case of a 5-year-old boy with a milder form of PMD and a PLP1 point deletion was described [26]. According to genotype-phenotype studies reported in literature a milder form of PMD developed from deletions of PLP1. Because the majority of PLP1 point mutations cause more severe dysmyelinating diseases than null mutations, Inoue speculated that at the PLP1 locus dominant-negative and loss-of-function alleles may result in different pathogenesis with distinct phenotypic consequences [16]. Although the patient we described showed a severe but stable phenotype, he present a exon 6 deletion in PLP1, confirming the variability in clinical expression of PMD and extending the knowledge of genotype-phenotype correlation. Since PLP1 deletions have been usually associated with milder form of PMD, we hypothised that possible modifier genes exist or other genetic mechanisms involved in etiopathogenesis of PMD could occur. Furthermore our patient in which we found a novel PLP1 deletion by NGS highlights the diagnostic utility of this technique to investigate PLP1 deletion as well as point mutations.
Acknowledgements
The authors thank the patient and his family for cooperation and support in this study. AMES Polidiagnostic Instrumental Centre supported this work within the research project: “Autismo: tra genetica ed Epigenetica”.
Authorship
G.S. enrolled the patients, performed and coordinated the laboratory experimental and data acquisition, and critical revision of manuscript, F.F.O. collected and provided clinical information, drafting of manuscript, R.R. carried out whole-exome sequencing, MLPA and data analysis; A.C. provided clinical information and critical revision of manuscript, R.R. carried out whole-exome sequencing, MLPA and interpretation of data, L.D.F. drafting of manuscript and critical revision of manuscript, G.C. interpretation of data, F.P. collected and provided clinical information, A.V. and R.M. literature review, A.F. study concept and design, analysis and interpretation of data, and critical revision of manuscript. ALL authors approved the final version of the article, including the authorship list.
References
- Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med, 2013;369:1502-11.
- Helbig KL, Farwell Hagman KD, Shinde DN, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016; 18: 898-905.
- Pajusalu S, Kahre T, Roomere H, et al. Large gene panel sequencing in clinical diagnostics-results from 501 consecutive cases. Clin Genet. 2018; 93: 78-83.
- Xie H, Feng H, Ji J, et al. Identification and functional study of novel PLP1 mutations in Chinese patients with Pelizaeus–Merzbacher disease. Brain Dev. 2015; 37: 797-802.
- Koeppen A H, and Robitaille Y. Pelizaeus-Merzbacher disease. J Neuropath Exp Neurol. 2002; 61: 747-759.
- Hoffman-Zacharska D, Mierzewska H, Mazurczak T, et al. Wrodzona postać choroby Pelizaeusa-Merzbachera jako wynik submikroskopowej duplikacji Xq21-22 obejmującej gen PLP1–opis przypadku. Neurol Dziecięca. 2013; 22: 73-77.
- Torii T, Miyamoto Y, Yamauchi J, et al. Pelizaeus-Merzbacher disease: Cellular pathogenesis and pharmacologic therapy. Pediatr Int. 2014; 56: 659-666.
- Garbern JY. Pelizaeus-Merzbacher disease: genetic and cellular pathogenesis. Cell Mol Life Sci. 2007; 64: 50-65.
- Mierzewska H, Jamroz E, Mazurczak T, et al. Pelizaeus-Merzbacher disease in patients with molecularly confirmed diagnosis. Folia Neuropathol. 2016 54: 59-65.
- Miyatake C, Koizumi S, Narazaki H, et al. Clinical pictures in Pelizaeus-Merzbacher disease: A report of a case. J Nippon Med Sch. 2015; 82: 74-75.
- Hobson GM, Garbern JY. Pelizaeus-Merzbacher disease, Pelizaeus-Merzbacher-like disease 1, and related hypomyelinating disorders. Semin Neurol. 2012; 32: 62-67.
- Lassuthová P, Zaliova M, Inoue K, et al. Three new PLP1 splicing mutations demonstrate pathogenic and phenotypic diversity of Pelizaeus-Merzbacher disease. J Child Neurol. 2014; 29: 924-931.
- Lu Y, Shimojima K, Sakuma T, et al. A novel PLP1 mutation F240L identified in a patient with connatal type Pelizaeus-Merzbacher disease. Hum. Genome Var. 2017; 4: 16044.
- Pavlidou E, Ramachandran V, Govender V, et al. A novel PLP1 mutation associated with optic nerve enlargement in two siblings with Pelizaeus–Merzbacher disease: A new MRI finding. Brain Dev. 2017; 39: 271-274.
- Masliah-Planchon J, Dupont C, Vartzelis G, et al. Insertion of an extra copy of Xq22. 2 into 1p36 results in functional duplication of the PLP1 gene in a girl with classical Pelizaeus-Merzbacher disease. BMC Med Genet. 2015; 16: 77.
- Inoue K, Osaka H, Thurston VC, et al. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet. 2002; 71: 838-853.
- Yiu EM, Farrell SA, Soman T. Classic Pelizaeus–Merzbacher disease in a girl with an unbalanced chromosomal translocation and functional duplication of PLP1. Mov Disord. 2009; 24: 2171-2172.
- Raskind WH, Williams CA, Hudson LD, et al. Complete deletion of the Proteolipid Protein Gene (PLP) in a family with X-linked Pelizaeus-Merzbacher disease. Am J Hum Genet. 1991; 49: 1355-1360.
- Torisu H, Iwaki A, Takeshita K, et al. Clinical and genetic characterization of a 2-year-old boy with complete PLP1 deletion. Brain Dev. 2012; 34: 852-856.
- Matsufuji M, Osaka H, Gotoh L, et al. Partial PLP1 deletion causing X-linked dominant spastic paraplegia type 2. Pediatr Neurol. 2013; 49: 477-481.
- Osaka H, Koizume S, Aoyama H, et al. Mild phenotype in Pelizaeus-Merzbacher disease caused by a PLP1-specific mutation. Brain Dev. 2010; 32: 703-707.
- Carvalho CMB, Bartnik M, Pehlivan D, et al. Evidence for disease penetrance relating to CNV size: Pelizaeus-Merzbacher disease and manifesting carriers with a familial 11 Mb duplication at Xq22. Clin Genet. 2012; 81: 532-541.
- Laššuthová P, Žaliová M, Inoue K, et al. Three new PLP1 splicing mutations demonstrate pathogenic and phenotypic diversity of pelizaeus-merzbacher disease. J Child Neurol. 2014; 29: 924-931.
- Warshawsky I, Chernova OB, Hübner CA, et al. Multiplex ligation-dependent probe amplification for rapid detection of proteolipid protein 1 gene duplications and deletions in affected males and carrier females with Pelizaeus-Merzbacher disease. Clin Chem. 2006;52:1267-1275.
- Brender T, Wallerstein D, Sum J, et al. Unusual presentation of Pelizaeus-Merzbacher disease: Female patient with deletion of the proteolipid protein 1 gene. Case Rep Genet. 2015: 453105.
- Shiihara T, Watanabe M, Moriyama K, et al. A novel PLP1 frameshift mutation causing a milder form of Pelizaeus–Merzbacher disease. Brain Dev. 2015; 37: 455-458.